pyhrf.sandbox.parcellation module
Hierarchical Agglomerative Clustering
These routines perform some hierarchical agglomerative clustering of some input data. Currently, only Ward’s algorithm
is implemented.
Authors : Vincent Michel, Bertrand Thirion, Alexandre Gramfort, Gael Varoquaux
Modified: Aina Frau
License: BSD 3 clause
-
class
pyhrf.sandbox.parcellation.AgglomerationTransform
Bases: object
-
class
pyhrf.sandbox.parcellation.BaseEstimator
Bases: object
-
class
pyhrf.sandbox.parcellation.ClusterMixin
Bases: object
-
pyhrf.sandbox.parcellation.FWHM(Y)
-
pyhrf.sandbox.parcellation.GLM_method(name, data0, ncond, dt=0.5, time_length=25.0, ndelays=0)
-
pyhrf.sandbox.parcellation.Memory(*args, **kwargs)
-
class
pyhrf.sandbox.parcellation.Ward(n_clusters=2, memory=None, connectivity=None, copy=True, n_components=None, compute_full_tree='auto', dist_type='uward', cov_type='spherical', save_history=False)
Bases: pyhrf.sandbox.parcellation.BaseEstimator, pyhrf.sandbox.parcellation.ClusterMixin
Ward hierarchical clustering: constructs a tree and cuts it.
| Parameters: |
- n_clusters (int or ndarray) – The number of clusters to find.
- connectivity (sparse matrix.) – Connectivity matrix. Defines for each sample the neighboring
samples following a given structure of the data.
Default is None, i.e, the hierarchical clustering algorithm is
unstructured.
- memory (Instance of joblib.Memory or str) – Used to cache the output of the computation of the tree.
By default, no caching is done. If a string is given, it is the
path to the caching directory.
- copy (bool) – Copy the connectivity matrix or work inplace.
- n_components (int (optional)) – The number of connected components in the graph defined by the connectivity matrix. If not set, it is estimated.
- compute_full_tree (bool or auto (optional)) – Stop early the construction of the tree at n_clusters. This is
useful to decrease computation time if the number of clusters is
not small compared to the number of samples. This option is
useful only when specifying a connectivity matrix. Note also that
when varying the number of cluster and using caching, it may
be advantageous to compute the full tree.
|
|---|
-
children_
array-like, shape = [n_nodes, 2] – List of the children of each nodes. Leaves of the tree do not appear.
-
labels_
array [n_samples] – cluster labels for each point
-
n_leaves_
int – Number of leaves in the hierarchical tree.
-
n_components_
sparse matrix. – The estimated number of connected components in the graph.
-
fit(X, var=None, act=None, var_ini=None, act_ini=None)
Fit the hierarchical clustering on the data
| Parameters: | X (array-like, shape = [n_samples, n_features]) – The samples a.k.a. observations. |
|---|
| Returns: | |
|---|
| Return type: | self |
|---|
-
class
pyhrf.sandbox.parcellation.WardAgglomeration(n_clusters=2, memory=None, connectivity=None, copy=True, n_components=None, compute_full_tree='auto', dist_type='uward', cov_type='spherical', save_history=False)
Bases: pyhrf.sandbox.parcellation.AgglomerationTransform, pyhrf.sandbox.parcellation.Ward
Feature agglomeration based on Ward hierarchical clustering
| Parameters: |
- n_clusters (int or ndarray) – The number of clusters.
- connectivity (sparse matrix) – connectivity matrix. Defines for each feature the neighboring
features following a given structure of the data.
Default is None, i.e, the hierarchical agglomeration algorithm is
unstructured.
- memory (Instance of joblib.Memory or str) – Used to cache the output of the computation of the tree.
By default, no caching is done. If a string is given, it is the
path to the caching directory.
- copy (bool) – Copy the connectivity matrix or work inplace.
- n_components (int (optional)) – The number of connected components in the graph defined by the
connectivity matrix. If not set, it is estimated.
- compute_full_tree (bool or auto (optional)) – Stop early the construction of the tree at n_clusters. This is
useful to decrease computation time if the number of clusters is
not small compared to the number of samples. This option is
useful only when specifying a connectivity matrix. Note also that
when varying the number of cluster and using caching, it may
be advantageous to compute the full tree.
- variance (array with variances of all samples (default: None)) – Injected in the calculation of the inertia.
- activation (level of activation detected (default: None)) – Used to weight voxels depending on level of activation in inertia computation.
A non-active voxel will not estimate the HRF correctly,
so features will not be correct either.
|
|---|
-
children_
array-like, shape = [n_nodes, 2] – List of the children of each nodes.
Leaves of the tree do not appear.
-
labels_
array [n_samples] – cluster labels for each point
-
n_leaves_
int – Number of leaves in the hierarchical tree.
-
fit(X, y=None, **params)
Fit the hierarchical clustering on the data
| Parameters: | X (array-like, shape = [n_samples, n_features]) – The data |
|---|
| Returns: | |
|---|
| Return type: | self |
|---|
-
pyhrf.sandbox.parcellation.align_parcellation(p1, p2, mask=None)
Align two parcellation p1 and p2 as the minimum
number of positions to remove in order to obtain equal partitions.
:returns: (p2 aligned to p1)
-
pyhrf.sandbox.parcellation.assert_parcellation_equal(p1, p2, mask=None, tol=0, tol_pos=None)
-
pyhrf.sandbox.parcellation.calculate_uncertainty(dm, g)
-
pyhrf.sandbox.parcellation.compute_fwhm(F, dt, a=0)
-
pyhrf.sandbox.parcellation.compute_hrf(method, my_glm, can, ndelays, i)
-
pyhrf.sandbox.parcellation.compute_mixt_dist(features, alphas, coord_row, coord_col, cluster_masks, moments, cov_type, res)
Within one given territory: bi-Gaussian mixture model with known posterior weights:
Estimation:
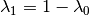 is the mean of posterior weights.
is the mean of posterior weights. 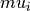 is estimated by weighted sample
mean and
is estimated by weighted sample
mean and 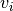 is estimated by weighted sample variance.
is estimated by weighted sample variance.
| Parameters: |
- features (np.array((nsamples, nfeatures), float)) – the feature to parcellate
- alphas (np.array(nsamples, float)) – confidence levels on the features -> identified to posterior weights of class activating in the GMM fit
- coord_row (list of int) – row candidates for merging
- coord_col (list of int) – col candidates for merging
- cluster_masks –
- moments –
- res –
|
|---|
-
pyhrf.sandbox.parcellation.compute_mixt_dist_skgmm(features, alphas, coord_row, coord_col, cluster_masks, moments, cov_type, res)
-
pyhrf.sandbox.parcellation.compute_uward_dist(m_1, m_2, coord_row, coord_col, variance, actlev, res)
Function computing Ward distance:
inertia = !!!!0
| Parameters: |
- m_1,m_2,coord_row,coord_col (clusters' parameters) –
- variance (uncertainty) –
- actlev (activation level) –
|
|---|
| Returns: |
- res (Ward distance)
- Modified (Aina Frau)
|
|---|
-
pyhrf.sandbox.parcellation.compute_uward_dist2(m_1, features, alphas, coord_row, coord_col, cluster_masks, res)
Function computing Ward distance:
In this case we are using the model-based definition to compute the inertia
| Parameters: |
- m_1,m_2,coord_row,coord_col (clusters' parameters) –
- variance (uncertainty) –
- actlev (activation level) –
|
|---|
| Returns: |
- res (Ward distance)
- Modified (Aina Frau)
|
|---|
-
pyhrf.sandbox.parcellation.feature_extraction(fmri_data, method, dt=0.5, time_length=25.0, ncond=1)
fmri_data (pyhrf.core.FmriData): single ROI fMRI data
-
pyhrf.sandbox.parcellation.generate_features(parcellation, act_labels, feat_levels, noise_var=0.0)
Generate noisy features with different levels across positions depending on parcellation and activation clusters.
| Parameters: |
- parcellation (np.ndarray of integers in [1, nb_parcels]) – the input parcellation
- act_labels (binary np.ndarray) – define which positions are active (1) and non-active (0)
- feat_levels (dict of (int : (array((n_features,), float), array(n_features), float)) -> (dict of (parcel_idx : (feat_lvl_inact, feat_lvl_act))) – map a parcel labels to feature levels in non-active and active-pos. Eg: {1: ([1., .5], [10., 15])} indicates
that features in parcel 1 have values [1., .5] in non-active positions (2 features per position) and value 10.
in active-positions
- noise_var (float>0) – variance of additive Gaussian noise
|
|---|
| Returns: | The simulated the features.
|
|---|
| Return type: | np.array((n_positions, n_features), float)
|
|---|
-
pyhrf.sandbox.parcellation.hc_get_heads(parents, copy=True)
Return the heads of the forest, as defined by parents
:param parents:
:type parents: array of integers
:param The parent structure defining the forest (ensemble of trees):
:param copy:
:type copy: boolean
:param If copy is False, the input ‘parents’ array is modified inplace:
| Returns: |
- heads (array of integers of same shape as parents)
- The indices in the ‘parents’ of the tree heads
|
|---|
-
pyhrf.sandbox.parcellation.hrf_canonical_derivatives(tr, oversampling=2.0, time_length=25.0)
-
pyhrf.sandbox.parcellation.informedGMM(features, alphas)
Given a set of features, parameters (mu, v, lambda), and alphas:
updates the parameters
WARNING: only works for nb features = 1
-
pyhrf.sandbox.parcellation.informedGMM_MV(fm, am, cov_type='spherical')
Given a set of multivariate features, parameters (mu, v, lambda), and alphas:
fit a GMM where posterior weights are known (alphas)
-
pyhrf.sandbox.parcellation.loglikelihood_computation(fm, mu0, v0, mu1, v1, a)
-
pyhrf.sandbox.parcellation.mixtp_to_str(mp)
-
pyhrf.sandbox.parcellation.norm2_bc(a, b)
broadcast the computation of ||a-b||^2
where size(a) = (m,n), size(b) = n
-
pyhrf.sandbox.parcellation.parcellation_hemodynamics(fmri_data, feature_extraction_method, parcellation_method, nb_clusters)
Perform a hemodynamic-driven parcellation on masked fMRI data
| Parameters: |
- fmri_data (-) – input fMRI data
- feature_extraction_method (-) – one of
‘glm_hderiv’, ‘glm_hdisp’ …
- parcellation_method (-) – one of
‘spatial_ward’, ‘spatial_ward_uncertainty’, …
|
|---|
| Returns: | parcellation array (numpy array of integers) with flatten
spatial axes
|
|---|
Examples #TODO
-
pyhrf.sandbox.parcellation.render_ward_tree(tree, fig_fn, leave_colors=None)
-
pyhrf.sandbox.parcellation.represent_features(features, labels, ampl, territories, t, fn)
Generate chart with features represented.
| Parameters: |
- features (-) – features to be represented
- labels (-) – territories
- ampl (-) – amplitude of the positions
|
|---|
| Returns: |
- the size of the spots depends on ampl,
and the color on labels
|
|---|
| Return type: | features represented in 2D
|
|---|
-
pyhrf.sandbox.parcellation.spatial_ward(features, graph, nb_clusters=0)
-
pyhrf.sandbox.parcellation.spatial_ward_sk(features, graph, nb_clusters=0)
-
pyhrf.sandbox.parcellation.spatial_ward_with_uncertainty(features, graph, variance, activation, var_ini=None, act_ini=None, nb_clusters=0, dist_type='uward', cov_type='spherical', save_history=False)
Parcellation the given features with the spatial Ward algorithm, taking into account uncertainty on features
(variance) and activation level:
- the greater the variance of a given sample, the lower its importance in the distance.
- the lower the activation level of a given sample, the lower its distance to any other sample.
| Parameters: |
- feature (np.ndarray) – observations to parcellate - size: (nsamples, nfeatures)
- graph (list of (list of PositionIndex)) – spatial dependency between positions
- variance (np.ndarray) – variance of features - size: (nsamples, nfeatures)
- activation (np.ndarray) – activation level associated with observation.
- size (int) – n samples
- var_ini –
- act_ini –
- nb_clusters (int) – number of clusters
- dist_type (str) – ward | mixt
|
|---|
-
pyhrf.sandbox.parcellation.squared_error(n, m)
-
pyhrf.sandbox.parcellation.ward_tree(X, connectivity=None, n_components=None, copy=True, n_clusters=None, var=None, act=None, var_ini=None, act_ini=None, dist_type='uward', cov_type='spherical', save_history=False)
Ward clustering based on a Feature matrix.
The inertia matrix uses a Heapq-based representation.
This is the structured version, that takes into account a some topological
structure between samples.
| Parameters: |
- X (array of shape (n_samples, n_features)) – feature matrix representing n_samples samples to be clustered
- connectivity (sparse matrix.) – connectivity matrix. Defines for each sample the neigbhoring samples
following a given structure of the data. The matrix is assumed to
be symmetric and only the upper triangular half is used.
Default is None, i.e, the Ward algorithm is unstructured.
- n_components (int (optional)) – Number of connected components. If None the number of connected
components is estimated from the connectivity matrix.
- copy (bool (optional)) – Make a copy of connectivity or work inplace. If connectivity
is not of LIL type there will be a copy in any case.
- n_clusters (int (optional)) – Stop early the construction of the tree at n_clusters. This is
useful to decrease computation time if the number of clusters is
not small compared to the number of samples. In this case, the
complete tree is not computed, thus the ‘children’ output is of
limited use, and the ‘parents’ output should rather be used.
This option is valid only when specifying a connectivity matrix.
- dist_type (str -> uward | mixt) –
|
|---|
| Returns: |
- children (2D array, shape (n_nodes, 2)) – list of the children of each nodes.
Leaves of the tree have empty list of children.
- n_components (sparse matrix.) – The number of connected components in the graph.
- n_leaves (int) – The number of leaves in the tree
- parents (1D array, shape (n_nodes, ) or None) – The parent of each node. Only returned when a connectivity matrix
is specified, elsewhere ‘None’ is returned.
- Modified (Aina Frau)
|
|---|
-
pyhrf.sandbox.parcellation.ward_tree_save(tree, output_dir, mask)


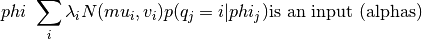
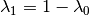 is the mean of posterior weights.
is the mean of posterior weights. 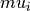 is estimated by weighted sample
mean and
is estimated by weighted sample
mean and 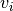 is estimated by weighted sample variance.
is estimated by weighted sample variance.